Estimating Uncertainty in the Black Hole Mass–Velocity Dispersion Relation#
Learning Objectives
Understand and implement bootstrap resampling to assess model uncertainty.
Practice using numpy to generate random samples.
Fit and interpret linear models.
Visualize parameter distributions to quantify fit variability.
Introduction#
The M-sigma relation, also known as the black hole mass-velocity dispersion relation, is an empirical correlation observed between the mass of supermassive black holes (\(M_{bh}\)) at the centers of galaxies and the velocity dispersion (\(\sigma\)) of stars in the bulges of those galaxies. This relationship suggests that there is a tight connection between the properties of galaxies and the central black holes they host. Specifically, galaxies with larger velocity dispersions tend to harbor more massive black holes at their centers.
The M-\(\sigma\) relation has profound implications for our understanding of galaxy formation and evolution, as well as the co-evolution of galaxies and their central black holes. It provides valuable insights into the mechanisms governing the growth and regulation of supermassive black holes, the role of feedback processes in galaxy evolution, and the relationship between the properties of galactic bulges and their central black holes.
Loading the data#
We’ll begin by loading some measurements of black hole mass (M_bh) and bulge velocity dispersion (sigma) for a set of nearby galaxies.
Important
Use the pandas library to load the m-sigma.txt file as a DataFrame (download here); it will read in with column names.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
df = pd.read_csv('m-sigma.txt')
df.head()
| Galaxy | M_bh | sigma | sigma_err | M_bh_err | |
|---|---|---|---|---|---|
| 0 | MW | 2950000.0 | 100 | 20 | 350000.0 |
| 1 | I1459 | 460000000.0 | 312 | 41 | 280000000.0 |
| 2 | N221 | 3900000.0 | 76 | 10 | 900000.0 |
| 3 | N3115 | 920000000.0 | 278 | 36 | 300000000.0 |
| 4 | N3379 | 135000000.0 | 201 | 26 | 73000000.0 |
Linear Regression#
Linear regression just means “let’s fit a line”! It’s a statistical method used to model the relationship between a dependent variable (often denoted as ‘y’) and one or more independent variables (often denoted as ‘x’). It assumes that the relationship between the variables can be described by a linear equation, such as a straight line in the case of simple linear regression. Like an inference task, the goal of linear regression is to find the best-fitting line that minimizes the differences between the observed values and the values predicted by the model.
For the simpliest case of a straight line, aka a first order polynomial, we can describe our model using two parameters: slope and intercept. In the case of the M-sigma relation, these fit parameters can be used to constrain our models of galaxy and black hole co-evolution.
How can we implement this in Python?#
There are many ways you can perform linear regression in Python! np.polyfit is a function in NumPy used for polynomial regression, which fits a polynomial curve to a set of data points. In the context of linear regression, np.polyfit can fit a polynomial of degree 1 (a straight line) to the given data points by minimizing the sum of squared errors between the observed and predicted values. This function returns the coefficients of the polynomial that best fits the data, allowing us to determine the slope and intercept of the regression line.
Beyond a Single Best-Fit Line: Representing Uncertainty in our Fit#
By now you should have produced a figure resembling the one below:
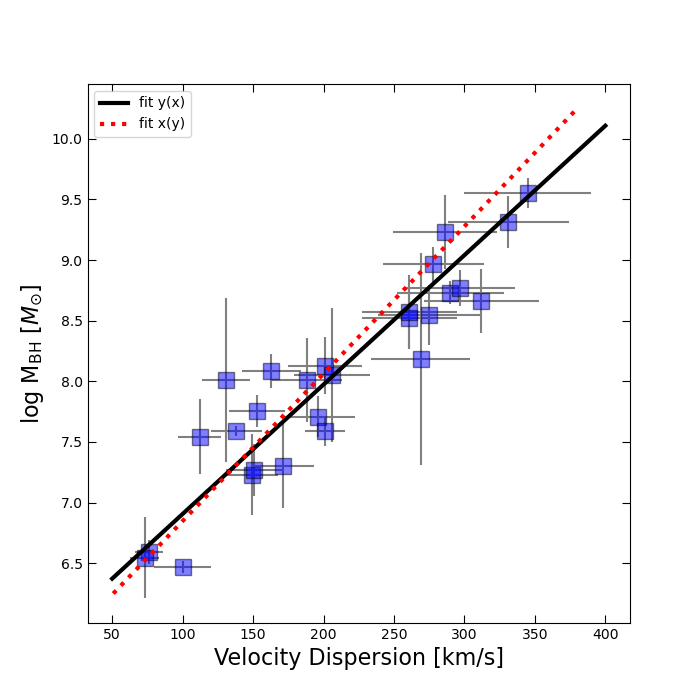
In our linear fits, we were only able to account for errors in one variable at a time. Standard least squares regression assumes that all uncertainty lies along the dependent variable (the y-axis). But in the M–\(\sigma\) relation, both quantities (black hole mass and velocity dispersion) have significant observational uncertainties. To explore this, we asked you to fit the relation in two directions: \(M_{\mathrm{bh}}(\sigma)\) and \(\sigma(M_{\mathrm{bh}})\).
As you probably noticed, these two fits give very different slopes! Neither is “more correct” than the other. Rather, the discrepancy reflects the fact that our data are noisy in both dimensions, and with limited data we cannot uniquely pin down the “true” relation. What we can say is that there is uncertainty in the fitted parameters of the M–\(\sigma\) relation.
Up to this point we’ve been thinking about measurement errors (the observational error bars on each galaxy’s \(M_{\mathrm{bh}}\) and \(\sigma\)). But there is a second kind of uncertainty that arises in astronomy: sample variance. If you repeated the experiment with a different set of galaxies, you might measure a slightly different relation. Since we cannot observe every galaxy in the universe, we need a way to estimate how much our finite sample might bias or vary our results.
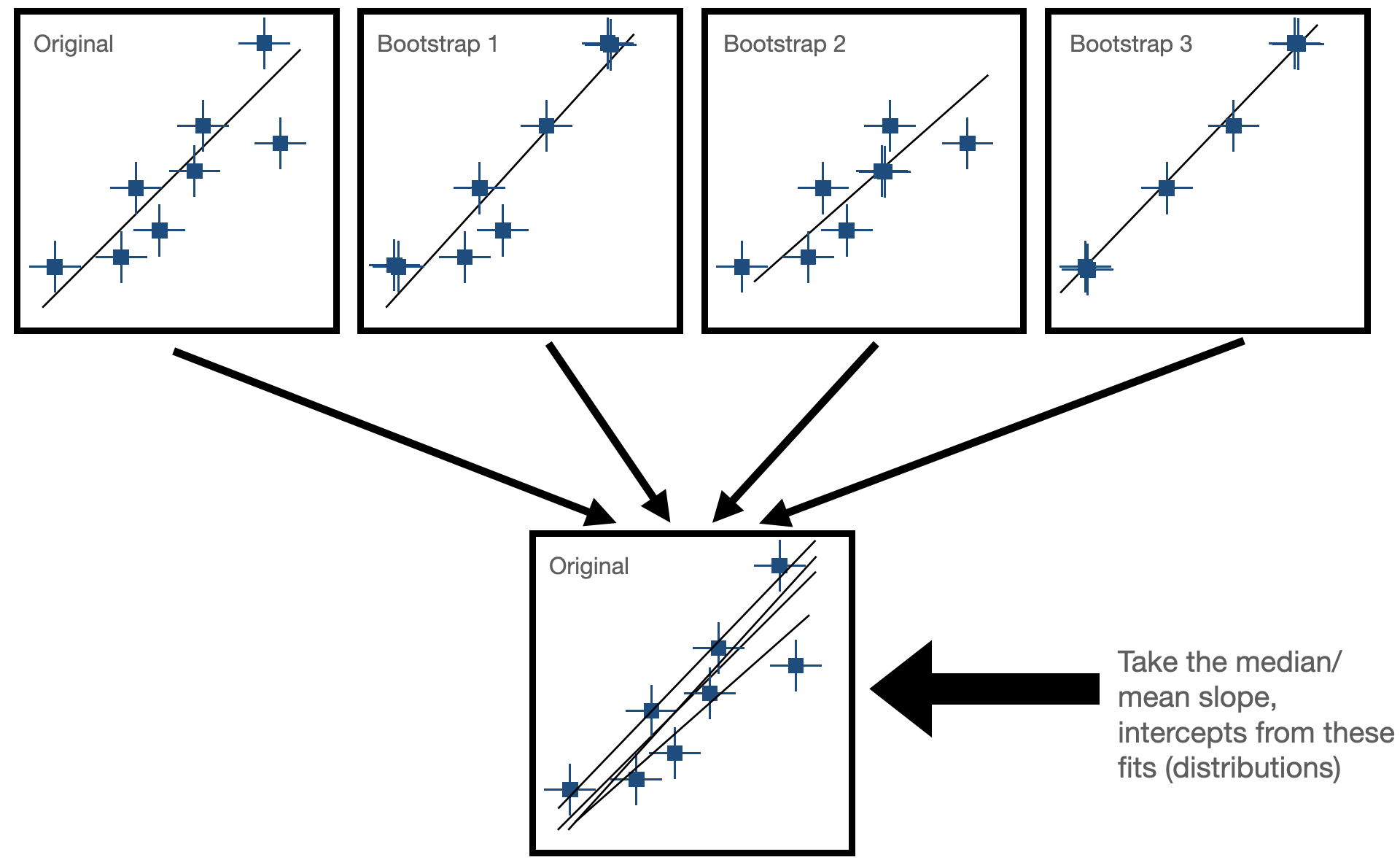
This is where bootstrap resampling comes in. Bootstrapping provides a way to simulate many alternative versions of our dataset and quantify how much the fitted parameters (slope and intercept) might change from sample to sample. Instead of quoting a single slope, we can quote a slope with an uncertainty range based on the distribution of slopes recovered from many bootstrap resamples.
Bootstrap Resampling#
Bootstrapping is a resampling technique used in statistics to estimate the sampling distribution of a statistic by repeatedly sampling with replacement from the observed data. It is particularly useful when the underlying population distribution is unknown or difficult to model. There are two general steps needed for bootstrap resampling:
Sample Creation: Bootstrapping starts with the creation of multiple bootstrap samples by randomly selecting observations from the original dataset with replacement. This means that each observation in the original dataset has the same chance of being selected for each bootstrap sample, and some observations may be selected multiple times while others may not be selected at all.
Statistical Estimation: After creating the bootstrap samples, the statistic of interest (e.g., mean, median, standard deviation, regression coefficient) is calculated for each bootstrap sample. This results in a distribution of the statistic across the bootstrap samples, known as the bootstrap distribution. In our case, this would be fitting our line to the data and seeing how much the paramters (slope and intercept) change for each fit. Now, we have some way of expressing uncertainty in our fitted parameters!
The goal for this next exercise is to use bootstrapping to estimate the uncertainty on our fit for the M-sigma relation!
Orthogonal Distance Regression and Bootstrapping Together#
So far, you have seen that fitting the M-\(\sigma\) relation while considering uncertainty is not straightforward:
Ordinary least squares can only incorporate uncertainties in one variable at a time.
Bootstrapping helps us capture how much our finite sample influences the fitted parameters.
But ideally, we would like to account for both sources of uncertainty at once: measurement errors in both \(M_{\mathrm{bh}}\) and \(\sigma\), and the sampling error from having only a finite number of galaxies.
Orthogonal Distance Regression (ODR), also known as total least squares, provides a way to fit a line that accounts for uncertainties in both x and y simultaneously. In Python, this is implemented in the scipy.odr module. ODR minimizes the perpendicular distance from each data point to the fit line, rather than only the vertical offset, and can be weighted by the uncertainties on both variables.
